Jul 6, 2020 | Elimu Informatics
The CDC has issued an RFI Concerning Personnel and the Retention of Next Generation Sequencing Data in Clinical and Public Health Laboratories. We (Elimu) would like to share our response to this RFI, as we feel that small changes in lab regulations have the potential to make much more sequencing data available for clinical decision support (CDS).
We encourage others to review the RFI and consider submitting their own or similar responses. Public comments will be used by the Clinical Laboratory Improvement Advisory Committee (CLIAC) for deliberation and possible recommendations about future changes to the Clinical Laboratory Improvement Amendments of 1988 (CLIA) regulations. CLIA plays a fundamental role in driving laboratory quality and consistency, and this RFI is particularly focused on factors that affect the quality of Next Generation Sequencing (NGS) data. Responses are due July 14.
————————————————
Elimu Informatics is a health informatics consulting and software development company, with a focus on genomics-EHR integration through CDS. To that end, we are developing a ‘Precision Genomics Integration Platform‘ that provides access to a patient’s full genomic testing results along with capabilities for the on-demand delivery of contextually relevant genomic findings and recommendations, via multiple EHR integration points. Primary components of the Platform include a FHIR-enabled genomic data server (GACS); Omnibus workflow and CDS engine; and Sapphire SMART-on-FHIR app assembler.
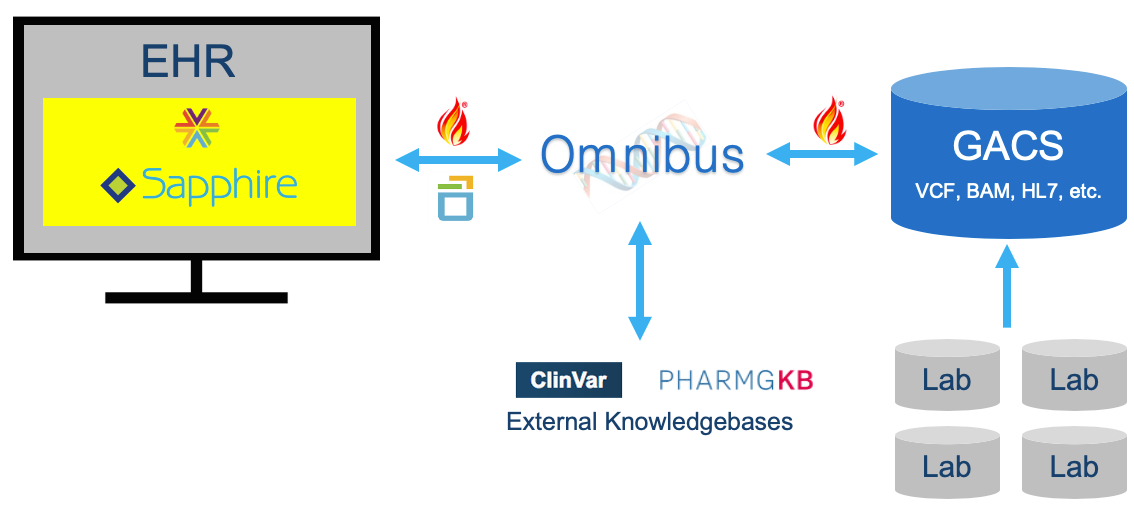
The biggest barrier we face in the development of this platform is access to a patient’s full genomic testing results. Several barriers exist. Some are technical. Some relate to the maturity of HL7 and GA4GH standards. But perhaps most relevant to CLIA, significant barriers relate to analytic capabilities of NGS and our state of genotype-phenotype knowledge. These latter barriers constrain genomics CDS by severely limiting the number of variants that labs are comfortable making available for clinical reasoning.
Issues around analytic validity (e.g. ability of a test to predict the presence of a variant) and clinical validity (e.g. ability of a variant to predict the presence of a disease) of variant identification and annotation pose significant barriers to Genomics-EHR integration. NGS is a complex process that involves determining a person’s DNA sequence and identifying how it differs from a reference DNA. The clinical significance of a variant is constantly evolving as our state of genotype-phenotype knowledge grows. Current analytic capabilities vary in their ability to differentiate true positive from false positive variants. Laboratories often perform confirmatory testing on key variants identified before including them in a report, which can limit the number of variants made available for clinical reasoning. For known variants, knowledge sources such as ClinVar provide a categorization with respect to clinical significance (e.g. ‘benign’, ‘likely pathogenic’) and review status (e.g. ‘single submitter, criteria provided’, ‘reviewed by expert panel’). Laboratories also consider these factors in determining which variants to report, again, at the potential expense of limiting what is returned. What is unclear, however, is whether the analytic and clinical validity considerations for laboratory reporting are the same as for CDS. For example, are there contexts in which a clinician would want to know about a pathogenic BRCA1 mutation, even if there is a small chance of false positive? Would a clinician ever want to know about a BRCA1 mutation only 90% likely to be pathogenic or where evidence of pathogenicity is scant? CDS engines are fundamentally different from polished lab reports in that the former have considerable capacity to deal with uncertainty, and tailor recommendations based on that uncertainty (e.g. by recommending confirmatory testing for a presumptive pathogenic variant).
As the CDC considers roles and responsibilities for personnel performing NGS, we suggest that different pipelines may warrant different skill sets. As the CDC considers examples of how NGS data files are used, we note that NGS data files are used extensively for clinical decision support. Examples include pharmacogenomic screening [Dolin RH, Boxwala A, Shalaby J. A Pharmacogenomics Clinical Decision Support Service Based on FHIR and CDS Hooks. Methods Inf Med. 2018 Dec;57(S 02):e115-e123] and other types of CDS-based precision medicine, such as ACMG secondary screening and variant reanalysis [HL7 Newsletter. “Point-of-Care Enabled Precision Medicine Service with GACS”. May, 2019. https://www.hl7.org/documentcenter/public/newsletters/HL7_NEWS_20190425.pdf]. We propose that CLIA could substantially advance Genomics-EHR integration by acknowledging that analytic and clinical validity considerations for a polished report delivered to an EHR from a lab are not necessarily the same as considerations applicable to the release of full genome data into a CDS engine, and that with careful consideration, the latter may be capable of reasoning against a wider set of variants.
————————————————
What are your thoughts on our thoughts? We’d love to hear from you!
